An Attention-Driven Hybrid Network for Survival Analysis of Tumorigenesis Patients Using Whole Slide Images
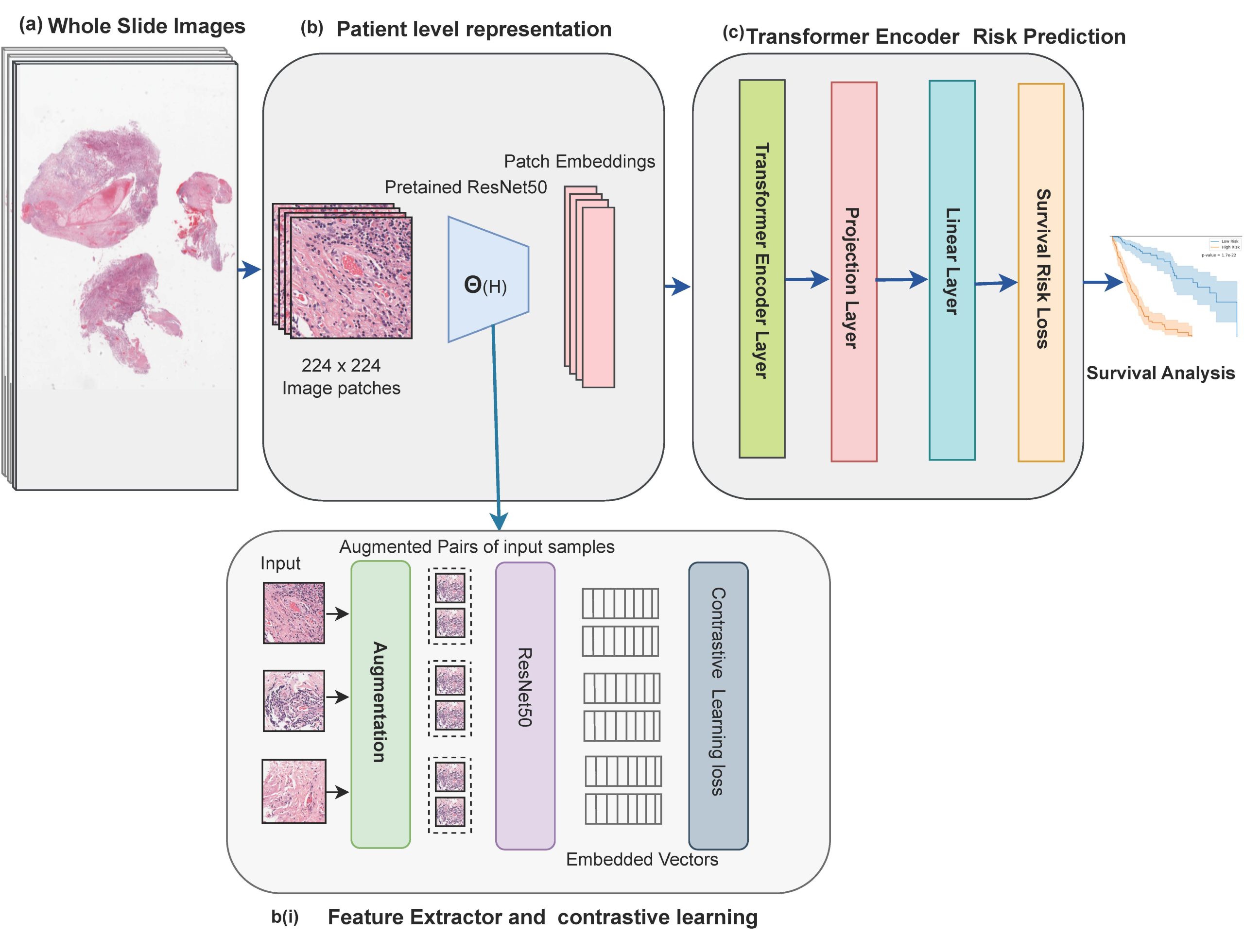
Survival analysis of cancer patients using Whole Slide Images (WSIs) is crucial in the field of medical statistics, as it helps identify key prognostic factors related to mortality and disease recurrence. However, extracting survival-relevant features from these images is a resource-intensive task and poses computational challenges. Instead of using the complete WSIs, most of the existing models rely on labor-intensive and time-consuming annotations by pathologists to extract features. Alternatively, many models take a selective approach by focusing on key patches from WSIs that potentially can miss important morphological details. Furthermore, most of these approaches employ Convolutional Neural Networks (CNNs) pre-trained on ImageNet to extract survival-related information. However, it’s important to note that there are significant differences in data distribution and domain-specific characteristics when comparing natural images to medical images. Our proposed methodology addresses these challenges by leveraging the advantages of contrastive learning and a transformer-based model. This architectural choice allows us to efficiently process and learn from the data, enabling us to capture intricate spatial relationships and contextual information within image patches. This capability is crucial for understanding the complex and heterogeneous nature of pathological tissues. Additionally, to make the most of the available information, we employ a strategy where we randomly select and utilize features extracted from 8,000 patches for each patient. This random selection process is carefully designed to encompass a diverse and relevant set of information from the patient’s dataset, ensuring a comprehensive representation of the input data within our transformer model. We evaluated the performance of our model using the TCGA-GBM and TCGA-LUSC datasets, and it achieved a concordance index (c-index) of 0.7892 and 0.744, respectively.
Video
Faculty
-
Dr. Muhammad Moazam FrazDr. Muhammad Moazam Fraz
-
Dr Zuhair Zafar
-
Dr Nazia Perwaiz
-
Dr Fahad Ahmad Satti
Students
-
Arshi Parvaiz
Publications
- S. Nasir, A. Parvaiz, M. M. Fraz. “Nuclei and glands instance segmentation in histology images: a narrative review”, In Artificial Intelligence Review 56 (8), 7909-7964 (2023) https://doi.org/10.1007/s10462-022-10372-5
- Parvaiz, M. A. Khalid, R. Zafar, H. Ameer, M. Ali, M. M. Fraz. “Vision Transformers in medical computer vision—A contemplative retrospection”, In Engineering Applications of Artificial Intelligence 122, 106126 (2023) https://doi.org/10.1016/j.engappai.2023.106126
- A. Parvaiz, E. S. Nasir, M. M. Fraz. “From Pixels to Prognosis: A Survey on AI-Driven Cancer Patient Survival Prediction Using Digital Histology Images”, In Journal of Imaging Informatics in Medicine, 1-24 (2024) https://doi.org/10.1007/s10278-024-01049-2
