Transforming Clinical Decision-Making: Predicting Antimicrobial Resistance from MALDI-TOF Data
Antimicrobial resistance (AMR) has become one of the biggest threats to modern medicine, with delays in diagnosis directly costing lives. Conventional Antimicrobial Susceptibility Tests (ASTs) can take up to three days and require highly trained microbiologists, which makes them inaccessible in many hospitals, especially in low-resource regions like Pakistan. To overcome this barrier, our project harnesses the power of deep learning to predict resistance profiles directly from MALDI-TOF mass spectrometry data within minutes. Instead of waiting for cultures to grow, doctors can obtain resistance predictions almost instantly, enabling faster clinical decision-making and improving patient outcomes.
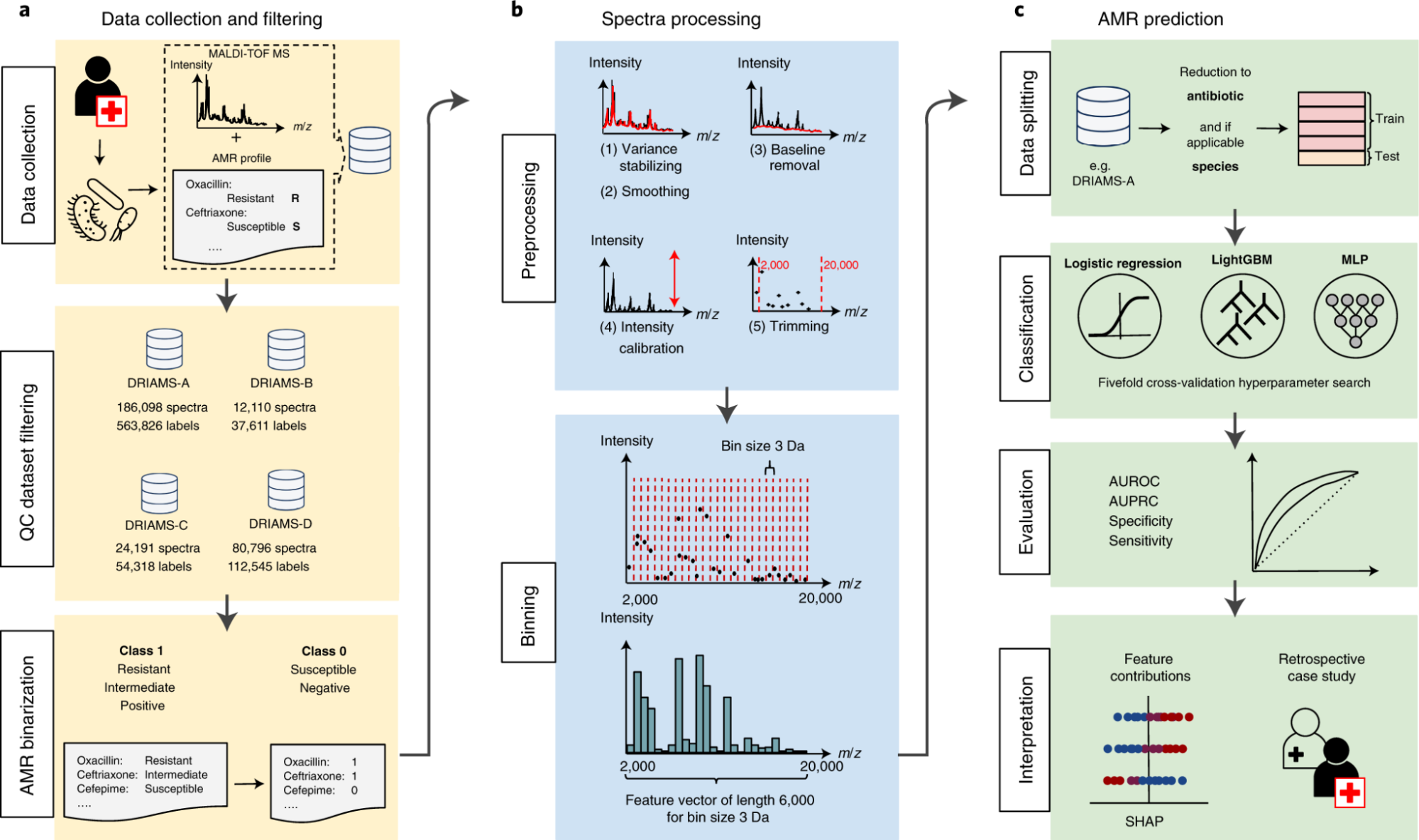
Our approach combines species-specific multi-label convolutional neural networks (CNNs), transfer learning, and continual learning strategies to create a scalable and adaptable pipeline. Starting with over 300,000 spectra from Swiss and German hospitals, the models are trained to recognize resistance signatures for critical pathogens such as Escherichia coli, Klebsiella pneumoniae, Staphylococcus aureus, and Pseudomonas aeruginosa. Through transfer learning, the models generalize across different hospital datasets, while continual learning mechanisms allow them to adapt to new bacterial strains and evolving antibiotic resistance patterns without catastrophic forgetting. This adaptability ensures that the system remains clinically relevant even as microbial landscapes shift over time.
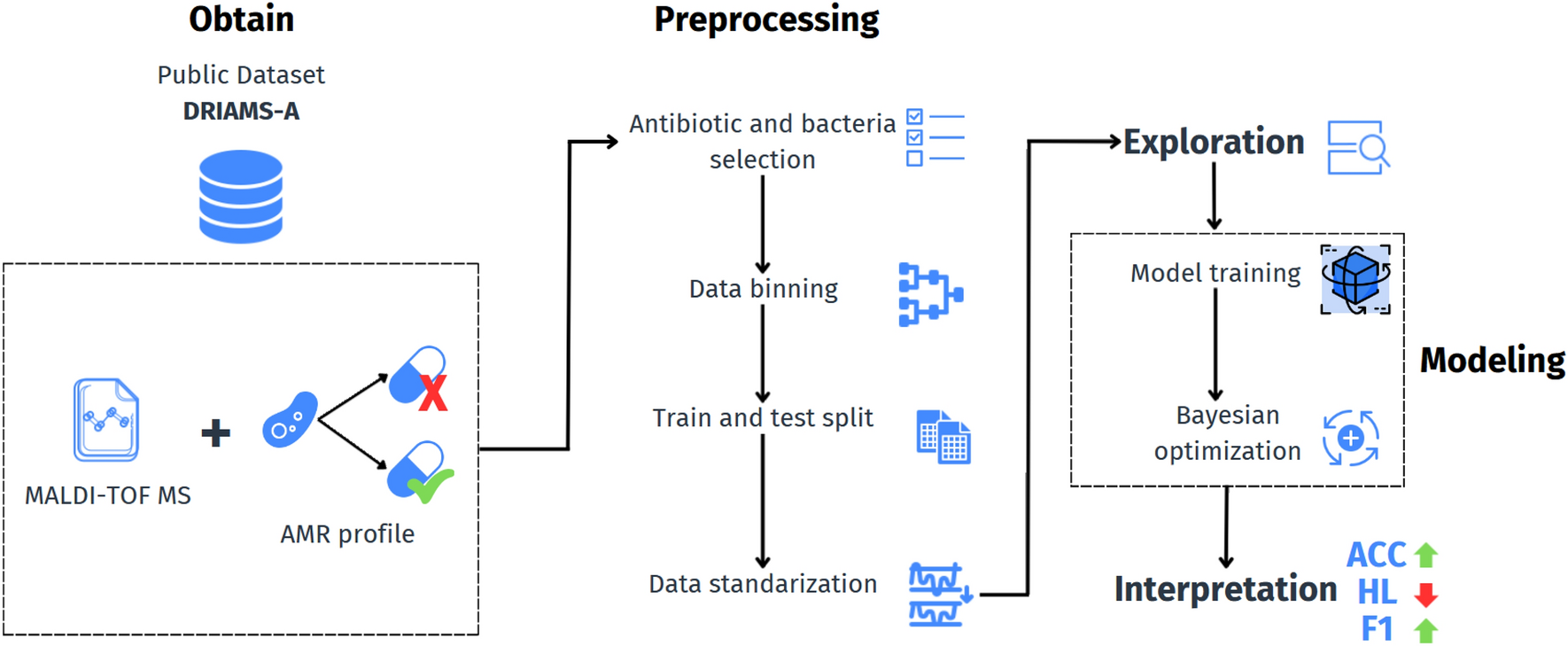
This project not only delivers high-performing models benchmarked against existing state-of-the-art systems but also lays the foundation for real-world deployment. By integrating the continual learning framework into a lightweight web API, the system can provide resistance predictions in real time, making it practical for use in hospitals worldwide. Beyond its direct clinical utility, the project demonstrates how AI can bridge the gap between cutting-edge microbiological research and frontline healthcare needs. It introduces a reproducible, generalizable, and future-proof pipeline that could transform how infections are diagnosed, ultimately reducing diagnostic delays, improving antibiotic stewardship, and strengthening global efforts to combat AMR.
Faculty
-
Dr. Muhammad Moazam FrazDr. Muhammad Moazam Fraz
-
Dr. Muhammad Naseer BajwaDr. Muhammad Naseer Bajwa
Students
-
Hasaan Hamid
-
Suman Kumari
-
Afra
