Cellular community detection for tissue phenotyping in digital histology images

Cellular community detection for tissue phenotyping in digital histology images
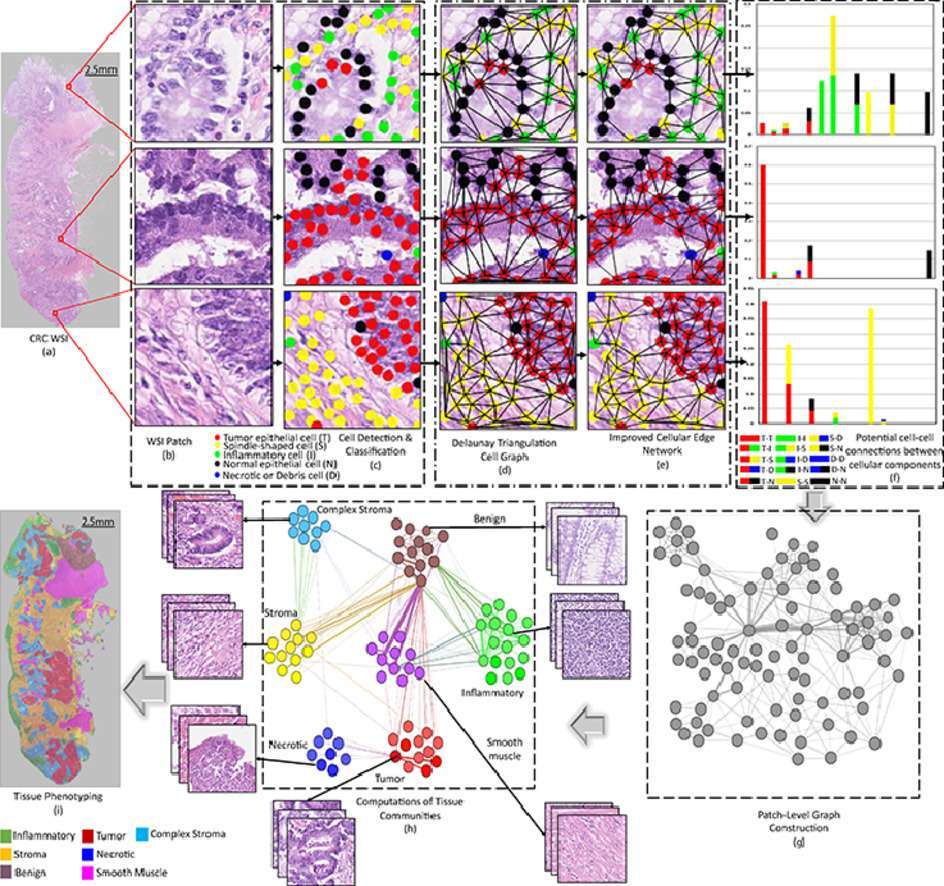
Classification of various types of tissue in cancer histology images based on the cellular compositions is an important step towards the development of computational pathology tools for systematic digital profiling of the spatial tumor microenvironment. Most existing methods for tissue phenotyping are limited to the classification of tumor and stroma and require large amount of annotated histology images which are often not available. In the current work, we pose the problem of identifying distinct tissue phenotypes as finding communities in cellular graphs or networks. First, we train a deep neural network for cell detection and classification into five distinct cellular components. Considering the detected nuclei as nodes, potential cell-cell connections are assigned using Delaunay triangulation resulting in a cell-level graph.
Based on this cell graph, a feature vector capturing potential cell-cell connection of different types of cells is computed. These feature vectors are used to construct a patch-level graph based on chi-square distance. We map patch-level nodes to the geometric space by representing each node as a vector of geodesic distances from other nodes in the network and iteratively drifting the patch nodes in the direction of positive density gradients towards maximum density regions. The proposed algorithm is evaluated on a publicly available dataset and another new large-scale dataset consisting of 280K patches of seven tissue phenotypes. The estimated communities have significant biological meanings as verified by the expert pathologists. A comparison with current state-of-the-art methods reveals significant performance improvement in tissue phenotyping.
Collaborators
-
Dr Muhammad Moazam FarazDr Muhammad Moazam Faraz
-
Prof Nasir M Rajpoot, TIA Lab, University of Warwick, UKProf Nasir M Rajpoot, TIA Lab, University of Warwick, UK
-
Dr Sajid Javed, Khalifa University Center for Autonomous Robotics System, UAEDr Sajid Javed, Khalifa University Center for Autonomous Robotics System, UAE
Selected Publications
- M.M. Fraz , S. A. Khurran, S. Graham, M. Shaban, Asif Loya, N.M. Rajpoot , “FABnet: Feature attention based network for simultaneous segmentation of microvessels and nerves in routine histology images of oral cancer”, Neural Computing and Applications, Vol. 20 , No. 1, Nov, 2019. IF: 4.66
- M.M.Fraz, M.Shaban, S.Graham, S.A.Khurram, N.M.Rajpoot , , “Uncertainty Driven Pooling Network for Microvessel Segmentation in Routine Histology Images”, Proceedings of the COMPAY workshop in 21st International Conference on Medical Image Computing and Computer Assisted Intervention, Sep, 2018, Granada , Spain.

